In a recent paper published in the American Journal of Human Genetics, postdoctoral research fellow Chelsea Lowther and graduate student Elise Valkanas co-led a large-scale study to systematically evaluate genome sequencing as a single test to displace the sequential application of karyotype, chromosomal microarray, and exome sequencing for the diagnostic assessment of autism spectrum disorder (ASD) and fetal structural anomalies (FSAs).
Leveraging data generated for 6,448 individuals as part of the SSC, Chelsea and Elise directly compared the diagnostic yields of three commonly used genetic technologies in the diagnostic assessment of ASD: chromosome microarray, exome sequencing, and genome sequencing. We established a standardized framework to identify diagnostic variants from genome and other sequencing (Figure 1).
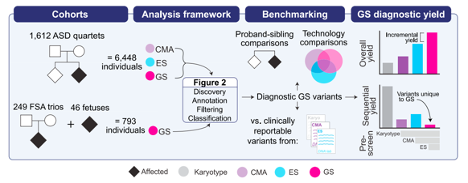
We demonstrated that our genome analysis framework could identify a diagnostic variant in 7.8% of ASD probands, almost 2-fold more than chromosomal microarray (4.3%) and 3-fold more than exome sequencing (2.7%). However, when we systematically captured copy-number variants (CNVs) from the exome data using a new tool developed by our lab (GATK-gCNV), the diagnostic yield of exome sequencing (7.4%) was brought much closer to, but did not surpass genome sequencing.
Next, we evaluated the performance of genome sequencing in 295 individuals with an FSA and found that genome sequencing could achieve an overall diagnostic yield of 46.1% in unselected FSAs, representing a 17.2% increased yield over karyotype, 14.1% over chromosomal microarray, and 4.1% over exome sequencing with CNV calling or 36.1% without. Overall, GS provided an added diagnostic yield of 0.4% and 0.8% beyond the combination of all three standard-of-care tests in ASD and FSAs, respectively. This corresponded to nine GS unique diagnostic variants, including sequence variants in exons not captured by ES, structural variants (SVs) inaccessible to existing standard-of-care tests, and SVs where the resolution of GS changed variant classification.
Overall, this study demonstrates that GS provides a significant increase in diagnostic yield over each individual test and a modest increase in yield beyond the combination of all three standard-of-care tests, thus warranting consideration as the first-tier diagnostic approach for the assessment of ASD and FSAs.
