The Talkowski Lab attended the AGBT precision health meeting in San Diego. As one of the founding AGBTph members and chair of the organizing committee, Mike helped plan the event and some lab members showcased their work.
We first spent the afternoon at a pre-meeting for the Fetal Genomics Consortium, which featured international consortia members and included Dr. Diana Bianchi, NICHD Director and former AGBTph OC member.
Then on to the main meeting. After an amazing opening session with Stephen Kingsmore, Daniel MacArthur, and Diana Bianchi giving keynotes, there was a Friday morning prenatal genomics session. This included talks by our FGC members Ron Wapner, Chris Barnett, and a featured abstract talk from our structural variant team lead Assistant professor Harrison Brand on our first presentation of comprehensive and high-resolution non-invasive prenatal screening from cell-free fetal DNA in maternal plasma. The talk highlighted the potential to capture all pathogenic variants in a fetal sample using a new non-invasive approach.
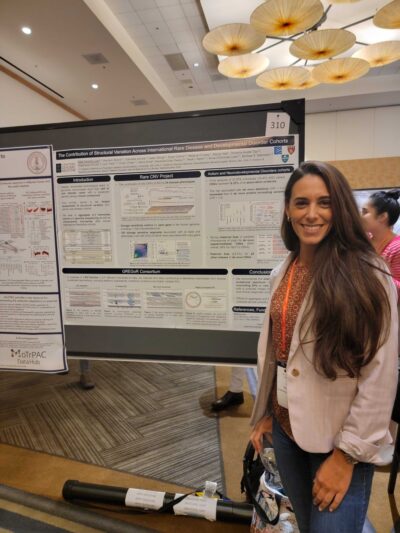
Postdoc Alba Sanchis-Juan then presented a poster investigating the contribution of structural variants to rare disease across exome and genome sequencing of international cohorts. She discussed preliminary analyses from nearly one million individuals to construct a genome-wide catalog of dosage sensitivity across 54 disorders and showed the overall contribution of coding and noncoding structural variants to rare disease cases. Efforts to aggregate and harmonize these analyses across cohorts are ongoing and have potential to impact both research and clinical diagnostic screening.
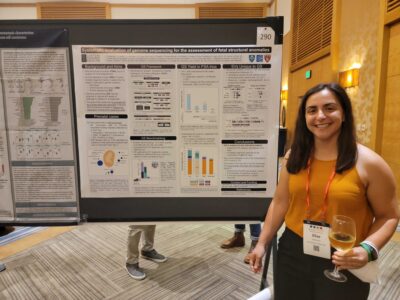
Elise Valkanas, a Harvard PhD student, also presented a poster detailing her work on using genome sequencing as a diagnostic test for the assessment of fetal structural anomalies. This work demonstrates that genome sequencing captures almost all variation discovered by other tests (karyotype, microarray, and exome sequencing) as well as additional, novel variants. For more on this work, see our preprint!
Finally, Katherine Chao from our gnomAD team at the Broad Institute presented a poster regional missense constraint in the human genome using gnomAD samples. She worked on this with our Harvard PhD student Lily Wang and one of the keynote speakers and CGM faculty members, Kaitlin Samocha.
Overall, it was a great meeting and wonderful to be together in San Diego for AGBTph.
- Dr. Harrison Brand
- Elise Valkanas and Poster
- Postdoc Abla Sanchis-Juan and Poster
- Dr. Mike Talkowski