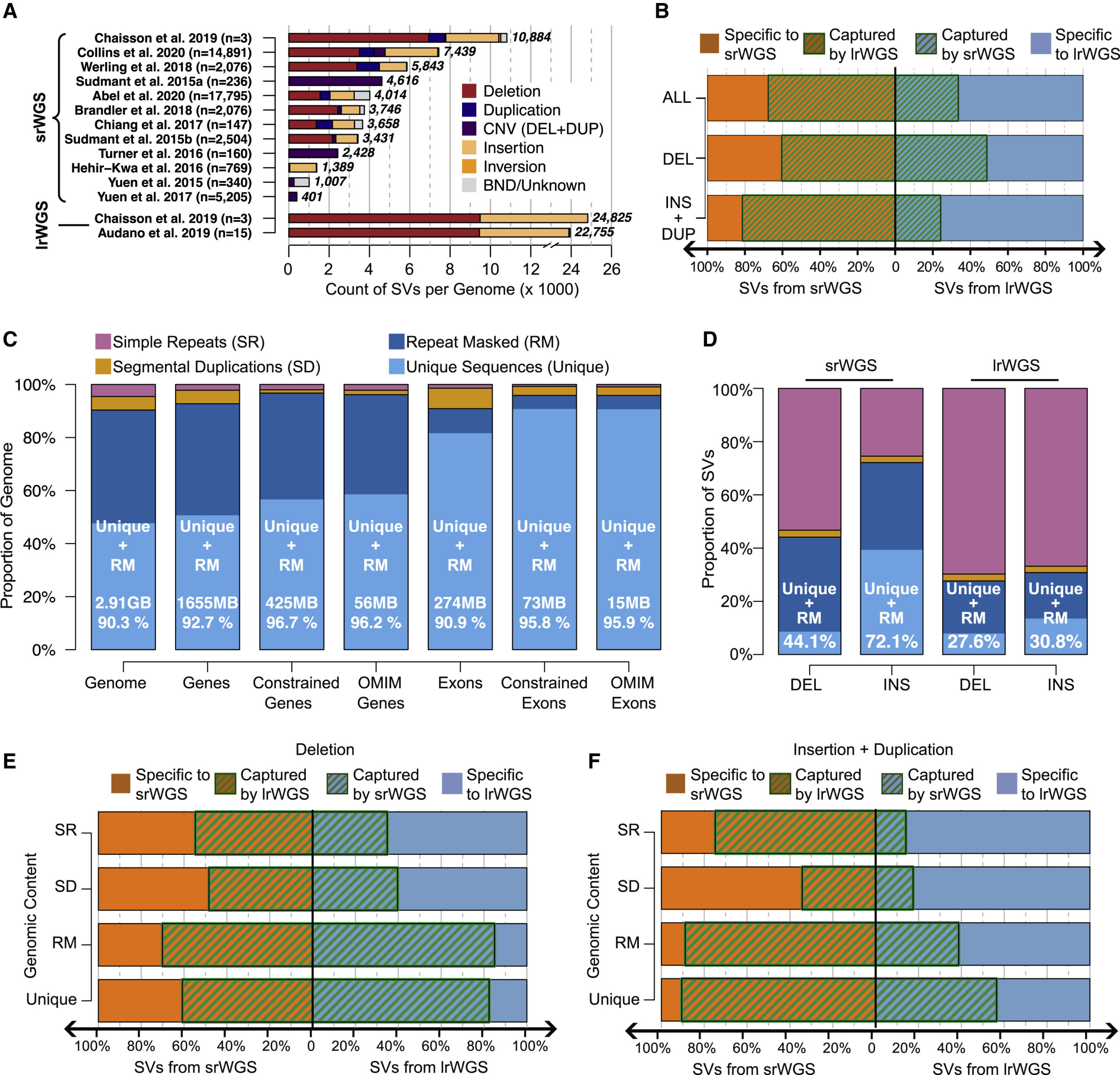
A structural variant catalog for All of Us
The Broad Institute's All of Us (AoU) Genome Center and All of Us Data and Research Center have led multiple aspects of variant discovery and data releases for this national biobank initiative. In June 2024, the laboratory of institute member and All of Us Co-I Mike Talkowski, including Emma Pierce-Hoffman, Mark Walker, Harrison Brand, Chris Whelan, [...]