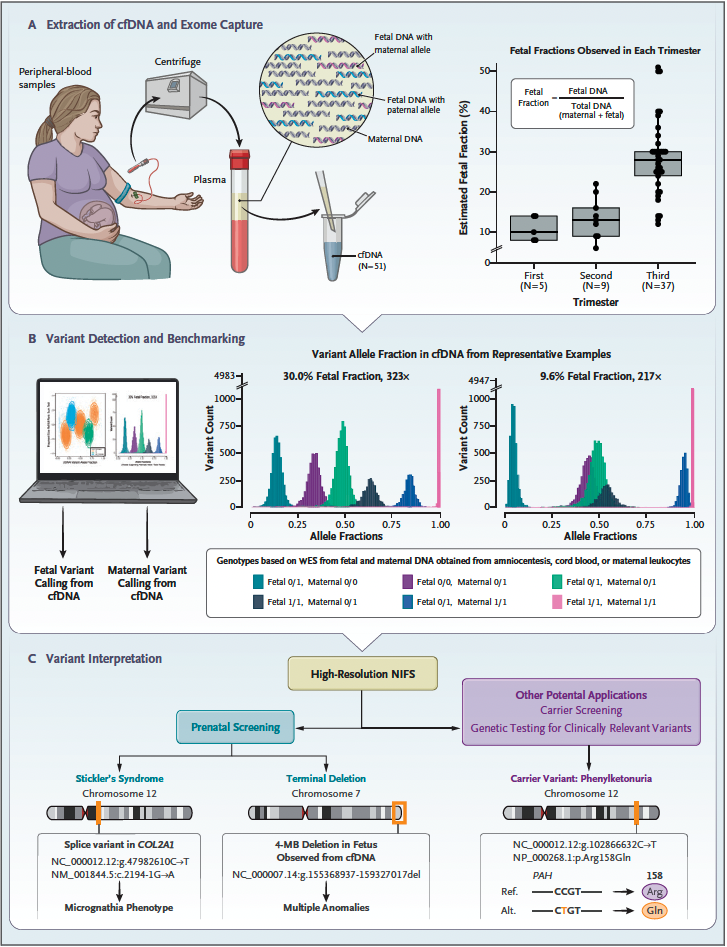
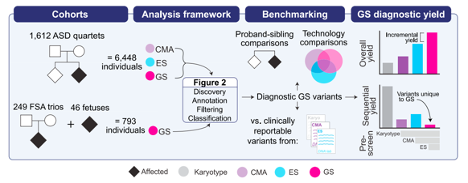
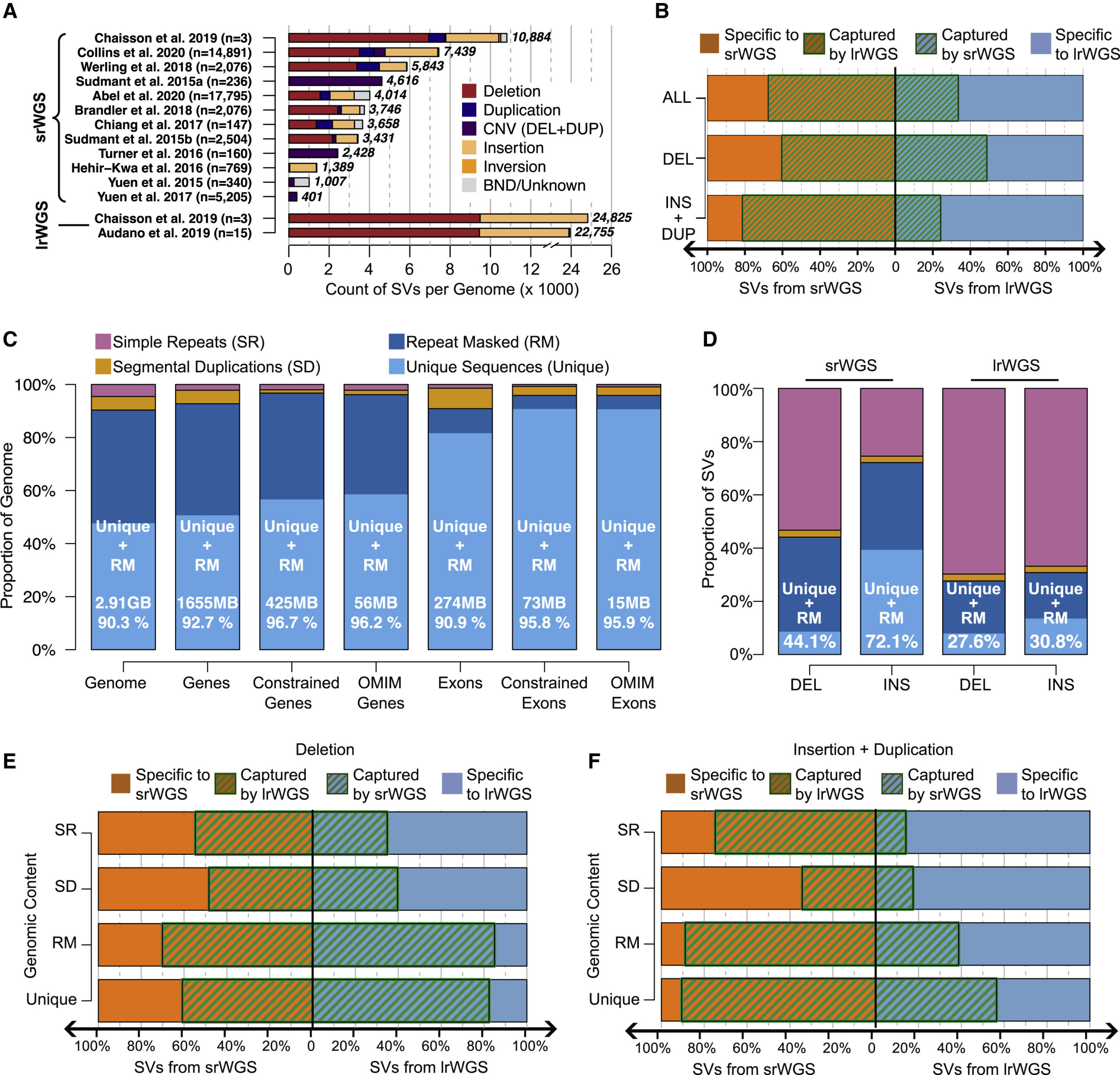
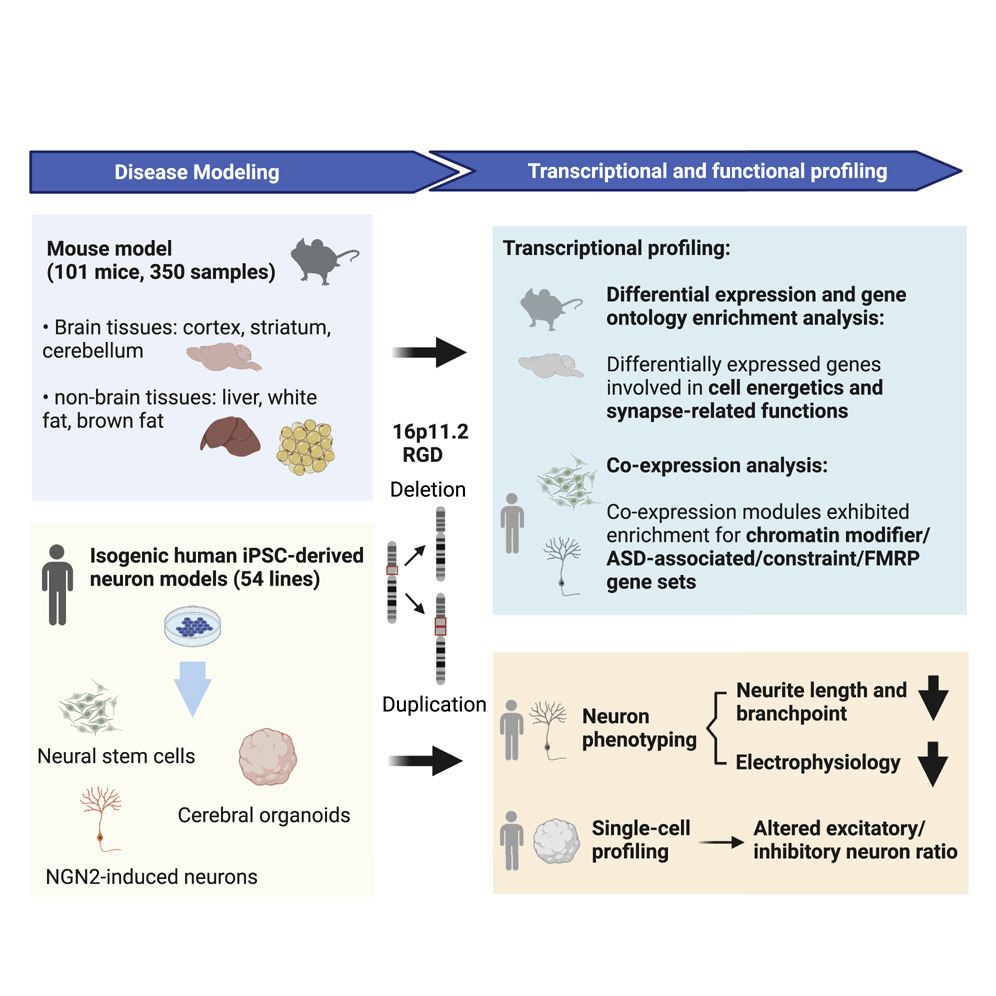
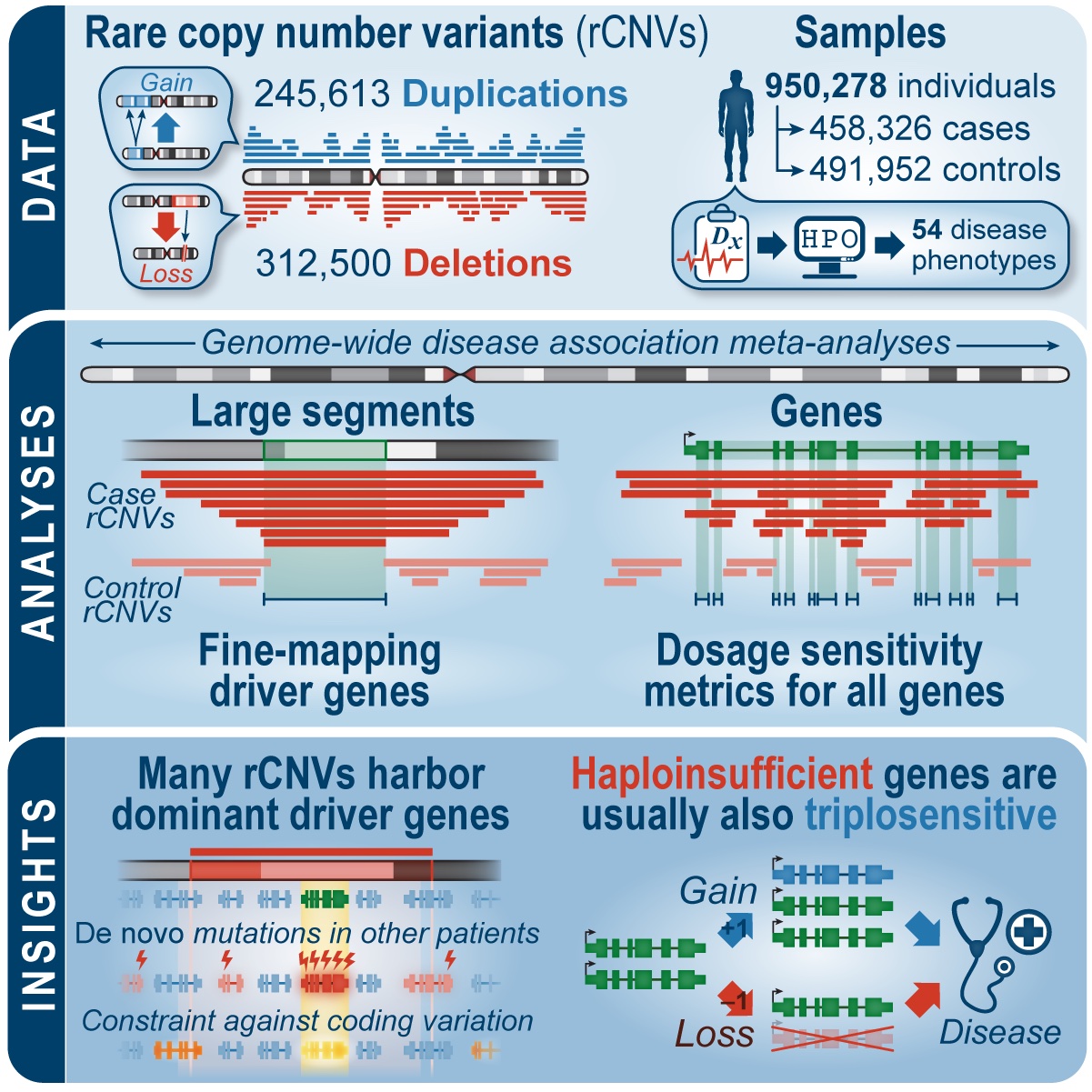
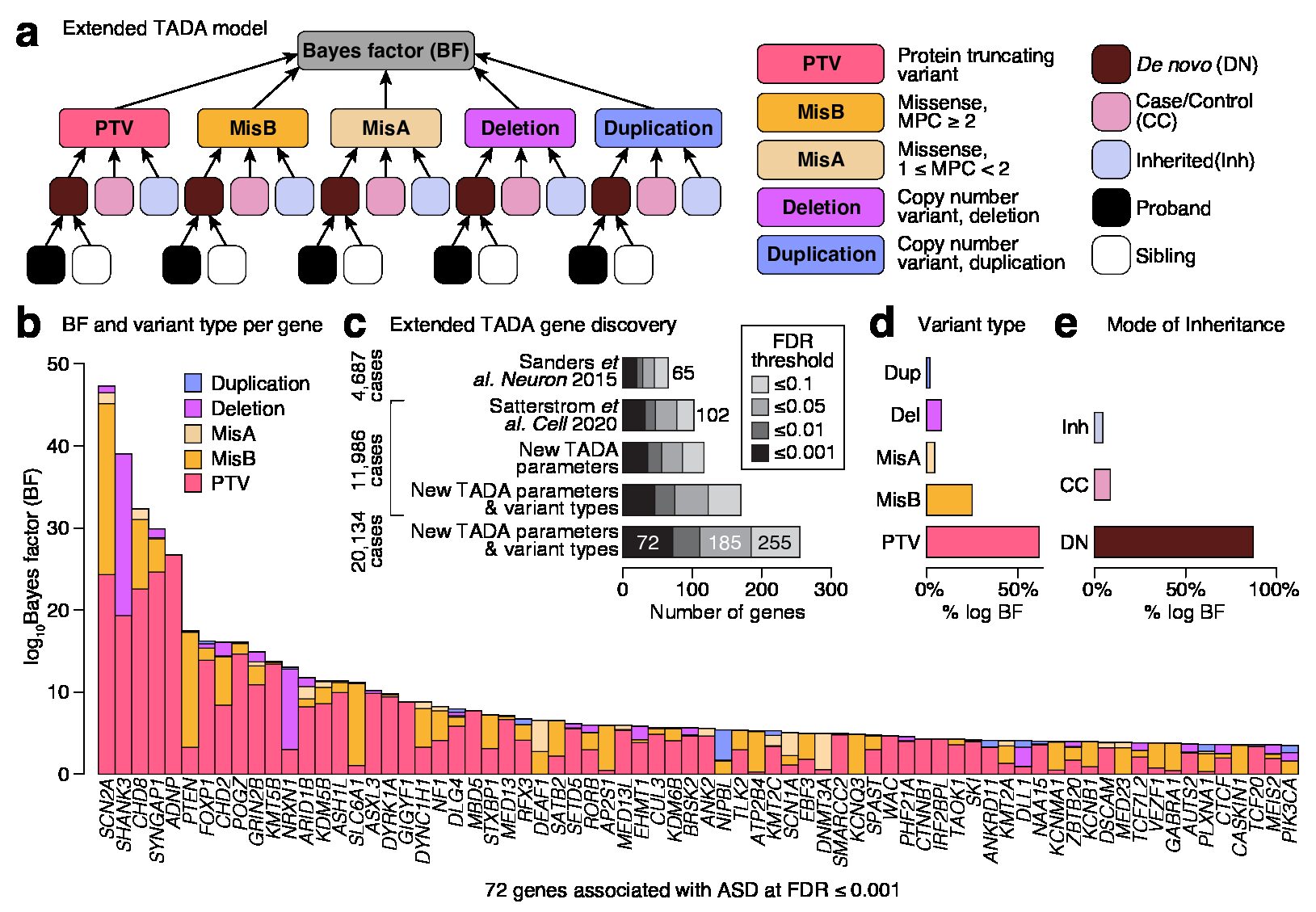
Nat Comm publication featured in the Genomics Research Spotlight NHGRI November 2024 newsletter!
Our recent article, published in Nature Communications in September 2024 and co-authored by Arthur Lee, postdoc fellow, and the Engle Lab at Boston Children's Hospital, was featured in the Genomics Research Spotlight of the NHGRI November 2024 issue of 'The Genomics Landscape'. You can read the full publication here!